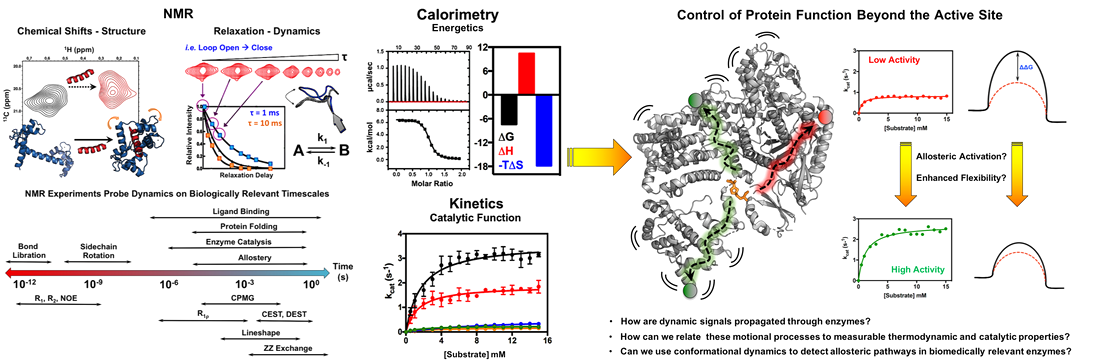
The Lisi laboratory utilizes solution NMR methods along with techniques in biochemistry, biophysics, and molecular biology to interrogate changes in protein structure and conformational motions that underlie function. We are especially focused on enzyme complexes, aiming to understand how biological events such as protein-protein interaction or the binding of allosteric effectors and drug-like molecules modulate functionally relevant protein dynamics, intra- and intermolecular signaling, and/or catalytic reactivity. There are several projects underway in the laboratory.
- Regulation of protein structure, dynamics, and interactions by local changes to cellular environments
|
We are particularly interested in determining the roles of two signaling proteins, macrophage migration inhibitory factor (MIF) and granulocyte-macrophage colony-stimulating factor (GM-CSF), in inflammatory responses connected to asthma, acute respiratory distress syndrome, and arthritis. Our investigations probe changes in the three-dimensional structures and multi-timescale conformational dynamics of these proteins, leveraging various biochemical stimuli (i.e. cellular redox conditions, mutations) to detect signatures of disease-related states. We aim for structural insight that will generate translational impact with potential for development of novel therapeutic approaches to inflammatory diseases.
|
- Pantouris, G.*; Khurana, L.; Ma, A.; Skeens, E.; Reiss, K.; Batista, V.S.; Lisi, G.P.*; Lolis, E.J.* Regulation of MIF Enzymatic Activity by an Allosteric Site at the Central Solvent Channel Cell Chem. Biol. 2020. 27. 740-750
- Chen, E.; Reiss, K.; Shah, D.; Manjula, R.; Allen, B.; Murphy, E.L.; Murphy, J.W.; Batista, V.S.; Bhandari, V.; Lolis, E.J.*; Lisi, G.P.* A Structurally Preserved Allosteric Site in the MIF Superfamily Affects Enzymatic Activity and CD74 Activation in D-dopachrome Tautomerase J. Biol. Chem. 2021. 297. 101061-101073
- Chen, E.; Reiss, K.; Shah, D.; Manjula, R.; Allen, B.; Murphy, E.L.; Murphy, J.W.; Batista, V.S.; Bhandari, V.; Lolis, E.J.*; Lisi, G.P.* A Structurally Preserved Allosteric Site in the MIF Superfamily Affects Enzymatic Activity and CD74 Activation in D-dopachrome Tautomerase J. Biol. Chem. 2021. 297. 101061-101073
- Protein-drug interactions
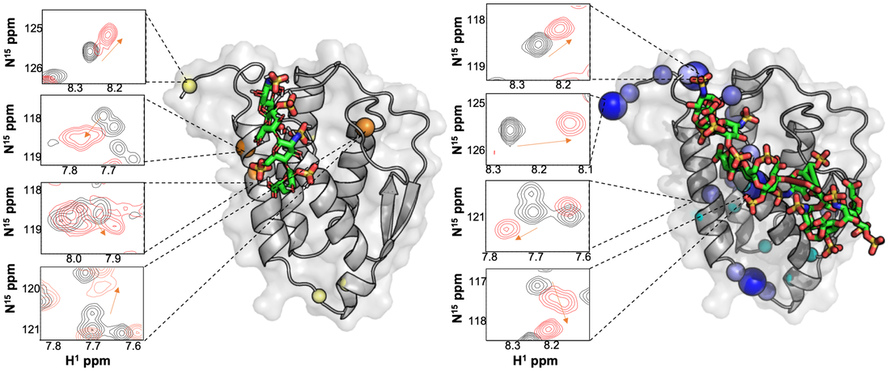
Sites of flexibility in proteins can modulate biological signaling, catalytic mechanisms, and interactions with binding partners. Targeting small molecules to these flexible regions is an avenue to develop novel points of functional control in MIF and GM-CSF, and to establish signatures of "free" (black) and "bound" (red) states.
- Cui, J.Y.; Zhang, F.; Nierzwicki, L.; Palermo, G.; Linhardt, R.J.; Lisi, G.P. Mapping the Structural and Dynamic Determinants of pH-sensitive Heparin Binding to Granulocyte Macrophage-colony Stimulating Factor Biochemistry. 2020. 59. 3541-3553
- Wang, J.; Shi, Y.; Reiss, K.; Maschietto, F.; Lolis, E.; Konigsberg, W.; Lisi, G.P.; Batista, V.S. Structural Insights into Binding of Remdesivir Triphosphate within the Replication-transcription Complex of SARS-CoV-2 Biochemistry. 2022. 61. 1966-1973
- Wang, J.; Shi, Y.; Reiss, K.; Maschietto, F.; Lolis, E.; Konigsberg, W.; Lisi, G.P.; Batista, V.S. Structural Insights into Binding of Remdesivir Triphosphate within the Replication-transcription Complex of SARS-CoV-2 Biochemistry. 2022. 61. 1966-1973
- "Dynamic Allostery" - Elucidating long-range signaling pathways in large enzyme complexes and machines
Molecular motions are critical to the optimization of enzyme scaffolds, occurring on timescales ranging from picoseconds-to-seconds that are often commensurate with steps of the catalytic cycle. These local fluctuations are known to propagate biological signals throughout the protein matrix, allowing enzyme complexes to synchronize the function of multiple domains. We aim to characterize these dynamic pathways so that we can a) pinpoint the chemical and biological activators of catalysis and b) identify points of functional control beyond the active site. These approaches are useful in elucidating chemical mechanisms and developing targeting strategies for disease-linked enzymes.
|
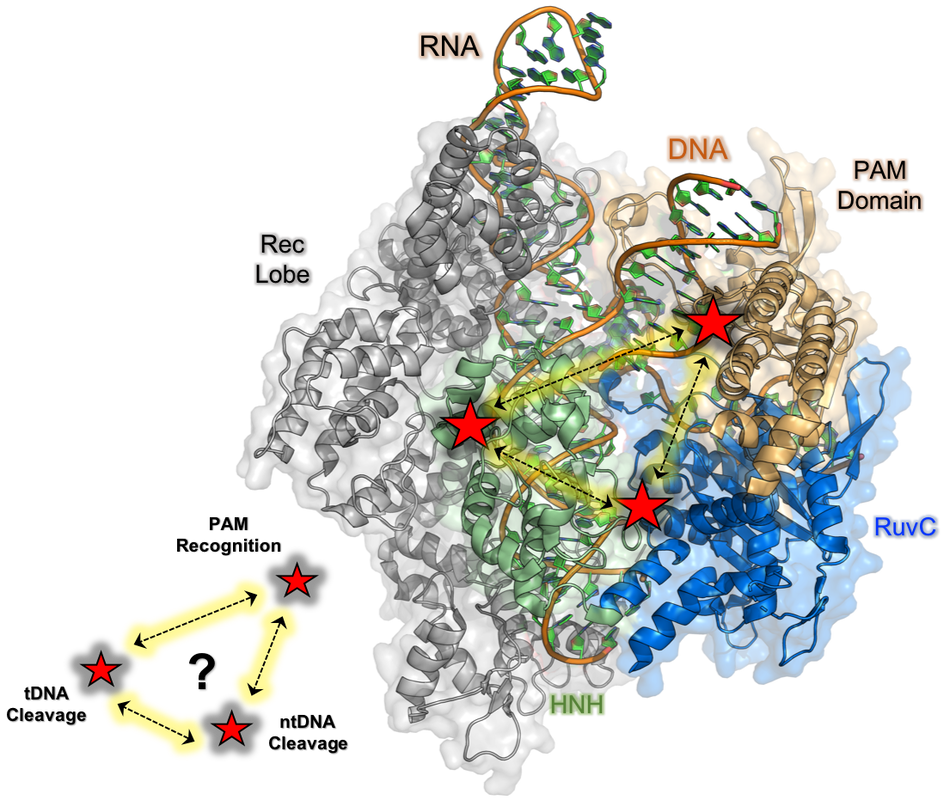
Our current work on the 1,350+ residue CRISPR-Cas9 endonuclease machinery aims to provide atomistic insight into the role of multi-timescale protein dynamics in transmitting biological (allosteric) information over large molecular distances to coordinate DNA cleavage. We investigate changes in the global structure and intrinsic dynamics of Cas9 subdomains in the presence of both its guide RNA and DNA substrate, allowing us to piece together the allosteric network of interactions that govern its function. Insight from our NMR studies are coupled to the state-of-the-art computational work of our collaborators (Victor Batista, Yale; Giulia Palermo, UC Riverside) that help to define the biochemical crosstalk controlling precise DNA cleavage. Insight from this work has potential to enhance spatial and temporal control over CRISPR-Cas9, which can improve its genome editing capabilities.
|
- East, K.W.; Newton, J.C.; Morzan, U.N.; Narkhede, Y.; Acharya, A.; Skeens, E.; Jogl, G.; Batista, V.S.; Palermo, G.*; Lisi, G.P.* Allosteric Motions of the CRISPR-Cas9 HNH Nuclease Probed by NMR and Molecular Dynamics J. Am. Chem. Soc. 2020. 142. 1348-1358
- Nierzwicki, L.; East, K.W.; Morzan, U.N.; Arantes, P.R.; Batista, V.S.; Lisi, G.P.*; Palermo, G.* Enhanced Specificity Mutations Perturb Allosteric Signaling in CRISPR-Cas9 eLife. 2021. 10. e73601 (LN and KWE contributed equally)
- Belato, H.B.; D'Ordine, A.M.; Nierzwicki, L.; Arantes, P.R.; Jogl, G.; Palermo, G.*; Lisi, G.P.* Structural and Dynamic Insight into the HNH Domain of Divergent Cas9 Species J. Struct. Biol. 2021. 214. 107814-107824
- Nierzwicki, L.; East, K.W.; Binz, J.; Hsu, R.V.; Arantes, P.R.; Ahsan, M.; Skeens, E.; Pacesa, M.; Jinek, M.; Lisi, G.P.*; Palermo, G.* Principles of Target DNA Cleavage and the Role of Mg2+ in the Catalysis of CRISPR-Cas9 Nature Catalysis. 2022. 5. 912-922
- Nierzwicki, L.; East, K.W.; Morzan, U.N.; Arantes, P.R.; Batista, V.S.; Lisi, G.P.*; Palermo, G.* Enhanced Specificity Mutations Perturb Allosteric Signaling in CRISPR-Cas9 eLife. 2021. 10. e73601 (LN and KWE contributed equally)
- Belato, H.B.; D'Ordine, A.M.; Nierzwicki, L.; Arantes, P.R.; Jogl, G.; Palermo, G.*; Lisi, G.P.* Structural and Dynamic Insight into the HNH Domain of Divergent Cas9 Species J. Struct. Biol. 2021. 214. 107814-107824
- Nierzwicki, L.; East, K.W.; Binz, J.; Hsu, R.V.; Arantes, P.R.; Ahsan, M.; Skeens, E.; Pacesa, M.; Jinek, M.; Lisi, G.P.*; Palermo, G.* Principles of Target DNA Cleavage and the Role of Mg2+ in the Catalysis of CRISPR-Cas9 Nature Catalysis. 2022. 5. 912-922
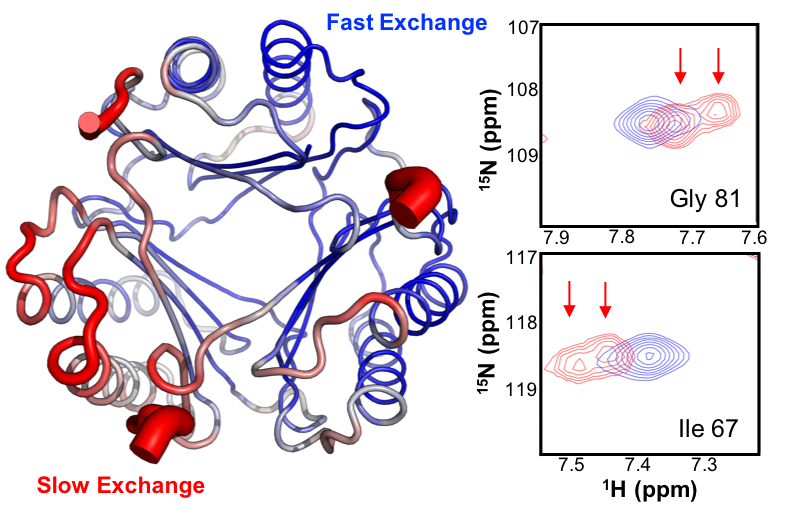
- "Latent Allostery" - Uncovering new or dormant functional sites in protein complexes
The ability to exert functional control over an enzyme from a region that is spatially distinct from its active site is a biological approach that is becoming popular in drug discovery. Allosteric phenomena are widely acknowledged in biochemistry, but it remains unclear exactly how or why allostery manifests itself in enzymes. The expansive and complex nature of protein structures suggest that an overwhelming percentage of allosteric regulatory sites remain undiscovered. We are interested in exploring unifying (structural and dynamic) principles of allostery that can facilitate the identification of these sites a priori and elucidate new biomedically relevant molecular mechanisms. We are currently using a family of bacterial enzymes, phosphoribosyl anthranilate isomerases (PRAIs), that catalyze the same chemical reaction despite significant structural, oligomeric, and mechanistic diversity, as a model to understand the biochemical and biophysical features most critical to allostery.
- Parkins, A.; Skeens, E.; McCallum, C.M.; Lisi, G.P.*; Pantouris, G.* The N-terminus of MIF Regulates the Dynamic Profile of Residues Involved in CD74 Activation Biophys. J. 2021. 120. 1-8
- Skeens, E.; Gadzuk-Shea, M.M.; Shah, D.; Bhandari, V.; Schweppe, D.K.; Berlow, R.B.*; Lisi, G.P.* Redox-dependent Structure and Dynamics of Macrophage Migration Inhibitory Factor Reveal Sites of Latent Allostery Structure. 2022. 30. 840-850
- Skeens, E.; Gadzuk-Shea, M.M.; Shah, D.; Bhandari, V.; Schweppe, D.K.; Berlow, R.B.*; Lisi, G.P.* Redox-dependent Structure and Dynamics of Macrophage Migration Inhibitory Factor Reveal Sites of Latent Allostery Structure. 2022. 30. 840-850
COLLABORATORS - We maintain collaborations with the following labs in our studies of enzyme biophysics
Prof. Ganesh Anand - Dept. of Chemistry, Penn State University - protein allostery via HDX mass spectrometry
Prof. Victor Batista - Dept. of Chemistry, Yale University - molecular simulations of CRISPR-Cas9 and MIF dynamics (R01 GM136815)
Dr. Frank Delaglio - Materials Measurement Lab, NIST - NMR sampling methods and spectral analysis
Prof. Gerwald Jogl - Dept. of Molecular Bio, Cell Bio & Biochemistry, Brown University - structure and function of CRISPR-Cas9
Prof. Megan Kizer - Dept. of Chemistry, Brown University - structural studies of protein-glycan interactions
Prof. Elias Lolis - Dept. of Pharmacology, Yale School of Medicine - structure and function of MIF and MIF-2
Dr. Sean Monaghan - Dept. of Surgery, Alpert Medical School at Brown - splicing of U1-70K in lung disease, clinical aspects of MIF/GMCSF
Dr. Alan Morrison - Providence VA Medical Center - structure and dynamics of the HuR RNA-binding protein (R01 HL163005)
Dr. Michael Ombrello - Translational Genetics and Genomics, NIAMS - molecular basis for inflammatory disease
Prof. Giulia Palermo - Dept. of Bioengineering, University of California, Riverside - CRISPR-Cas9 allostery in silico (R01 GM136815)
Prof. Georgios Pantouris - Dept. of Chemistry, University of the Pacific - structure and function of MIF
Prof. Brenda Rubenstein - Dept. of Chemistry, Brown University - experimental and computational studies of lowly-populated structures
Prof. Devin Schweppe - Dept. of Genome Sciences, University of Washington - mass spec studies of redox-driven protein modifications
Dr. Jimin Wang - Dept. of Molecular Biophysics & Biochemistry, Yale University - structure and dynamics of SARS-CoV-2 proteins
Prof. Ganesh Anand - Dept. of Chemistry, Penn State University - protein allostery via HDX mass spectrometry
Prof. Victor Batista - Dept. of Chemistry, Yale University - molecular simulations of CRISPR-Cas9 and MIF dynamics (R01 GM136815)
Dr. Frank Delaglio - Materials Measurement Lab, NIST - NMR sampling methods and spectral analysis
Prof. Gerwald Jogl - Dept. of Molecular Bio, Cell Bio & Biochemistry, Brown University - structure and function of CRISPR-Cas9
Prof. Megan Kizer - Dept. of Chemistry, Brown University - structural studies of protein-glycan interactions
Prof. Elias Lolis - Dept. of Pharmacology, Yale School of Medicine - structure and function of MIF and MIF-2
Dr. Sean Monaghan - Dept. of Surgery, Alpert Medical School at Brown - splicing of U1-70K in lung disease, clinical aspects of MIF/GMCSF
Dr. Alan Morrison - Providence VA Medical Center - structure and dynamics of the HuR RNA-binding protein (R01 HL163005)
Dr. Michael Ombrello - Translational Genetics and Genomics, NIAMS - molecular basis for inflammatory disease
Prof. Giulia Palermo - Dept. of Bioengineering, University of California, Riverside - CRISPR-Cas9 allostery in silico (R01 GM136815)
Prof. Georgios Pantouris - Dept. of Chemistry, University of the Pacific - structure and function of MIF
Prof. Brenda Rubenstein - Dept. of Chemistry, Brown University - experimental and computational studies of lowly-populated structures
Prof. Devin Schweppe - Dept. of Genome Sciences, University of Washington - mass spec studies of redox-driven protein modifications
Dr. Jimin Wang - Dept. of Molecular Biophysics & Biochemistry, Yale University - structure and dynamics of SARS-CoV-2 proteins